import pyeodh
import xarray as xr
import rioxarray
import os
from pathlib import Path
import requests
import threadingUser Example: Temporal Mosaics
Description & purpose: This Notebook introduces the science behind a user example looking to create temporal best pixel temporal mosaics using the Sentinel 2 ARD dataset. It also introduces the EOAP that has been created to run a scaled workflow on EODH.
Author(s): Alastair Graham, Dusan Figala
Date created: 2024-11-08
Date last modified: 2025-03-11
Licence: This file is licensed under Creative Commons Attribution-ShareAlike 4.0 International. Any included code is released using the BSD-2-Clause license.
Copyright (c) , All rights reserved.
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer. Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS “AS IS” AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
Background
In this example we are required to create a dataset that is cloud-free and temporally mosaiced. The input dataset is to be the CEDA Sentinel 2 ARD image data.
Cloud-free temporal mosaics of optical satellite images are composites created by combining multiple satellite acquisitions over a specific time period to generate a seamless and cloud-free view of a region. Optical satellite imagery, such as data from Sentinel-2, is often obstructed by clouds and atmospheric conditions, limiting the usability of individual acquisitions. Temporal mosaics overcome this challenge by selecting the clearest pixels from a time series of images, typically based on cloud masking algorithms or quality indices. These mosaics preserve spatial and spectral information while minimizing data gaps caused by clouds. Additional methods of data creation include using cloud-masks to remove clouds and/or shadow before the mosaicing step.
A user would want a cloud-free temporal mosaic of Sentinel-2 data to ensure consistent, high-quality imagery for analyzing surface features and environmental changes without the interference of clouds or shadows. Sentinel-2, part of the European Space Agency’s Copernicus program, provides high-resolution optical imagery across 13 spectral bands, making it ideal for applications in agriculture, forestry, urban planning, and climate monitoring. By creating a cloud-free mosaic, users can integrate multiple acquisitions into a single, seamless dataset that eliminates gaps caused by clouds and ensures better temporal and spatial coverage.
Analysis Ready Data (ARD) refers to satellite data that has been pre-processed and formatted to a standardized level, enabling users to perform analysis without needing to conduct significant additional preprocessing. ARD is designed to be easily accessible, interoperable, and suitable for a wide range of scientific and operational applications. The pre-processing typically includes geometric correction, radiometric calibration and atmospheric correction (removing the effects of the atmosphere on reflectance). It may also include cloud masking. The aim of creating ARD is to ensure data is consistent across time and space, and minimise the technical barriers for users, allowing them to focus on extracting insights rather than preparing raw data. By eliminating much of the preprocessing workload, ARD democratises the use of satellite data, allowing non-specialists and researchers to use the data more efficiently and reliably.
Such temporal mosaics are particularly valuable for applications requiring reliable, large-area analysis. For example, in agriculture, a cloud-free mosaic allows precise monitoring of crop health, phenological changes, and yield estimation by preserving uninterrupted vegetation indices. In forestry, it supports accurate assessments of deforestation, canopy density, and biomass. For urban planning and disaster management, cloud-free mosaics provide unobstructed views of land use changes, infrastructure, and affected regions, critical for decision-making.
The Scientific Process
Before creating a workflow on the EODH platform it was important to step through the scientific process to make sure that the correct tools and data were being considered.
To create the cloud-free outputs we use the cloud mask supplied with the ARD dataset. The information held in the cloud mask layer is used to block out the existing areas of cloud, and is applied to every band.
The temporal mosaic is created using a command line set of tools called pktools. Although a slightly older set of tools it is incredibly versatile, and being command-line first lends itself to the EOAP/CWl way of working. One of the tools, pkcomposite, is used to generate a median image from all input images over a one month period.
Processing steps
- pyeodh search of sentinel2_ard STAC (inputs are an AOI polygon (we need this later) and a date range)
- STEP 1: for each band in each returned image apply the cloud mask to remove cloud pixels from the data
- this results in a multibanded image that is cloud-masked
- need to retain information on original image capture date
- output should be a temporary STAC catalog
- STEP 2: for each cloud-masked image in a given month create band composites cropped to the AOI
- e.g.
pkcomposite -i in_A.tif -i in_B.tif -i in_C.tif -o out_b1.tif -cr median -b 0 -e aoi.geojson - need to be able to supply all images, the aoi polygon, and the composite method (e.g. median)
- output should be a STAC catalog of spatially cropped single-band composites for each month (e.g. for a three month period and 4 band input data there should be 3x Band1 outputs, 3x Band2 outputs, 3x Band3 outputs, 3x Band4 outputs)
- e.g.
The following tutorial demonstrates how to process data held in the Sentinel S2 ARD STAC.
Scientific method
First we need to import the required packages.
# set the area of interest
thetford_aoi = {
"coordinates": [
[
[0.08905898091569497, 52.69722175598818],
[0.08905898091569497, 52.15527412683906],
[0.9565339502005088, 52.15527412683906],
[0.9565339502005088, 52.69722175598818],
[0.08905898091569497, 52.69722175598818],
]
],
"type": "Polygon",
}Next we need to connect to the resource catalogue and undertake a search of the sentinel2_ard catalogue for the AOI location within a specific date range
# connect to
rc = pyeodh.Client(
base_url="https://staging.eodatahub.org.uk"
).get_catalog_service()
items = rc.search(
collections=["sentinel2_ard"],
catalog_paths=["supported-datasets/catalogs/ceda-stac-catalogue"],
intersects=thetford_aoi,
query=[
"start_datetime>=2023-04-01",
"end_datetime<=2023-06-30",
],
limit=1,
)Next, for each item open the cog, cloud and valid .tif assets and download them if needed.
for item in items.get_limited()[0:1]:
cloud_href = item.assets["cloud"].href
valid_href = item.assets["valid_pixels"].href
cog_href = item.assets["cog"].href
item
print(item.id, cloud_href, valid_href, cog_href, sep="\n")
valid = rioxarray.open_rasterio(
valid_href,
chunks=True,
)
cloud = rioxarray.open_rasterio(
cloud_href,
chunks=True,
)
# Check if cog file exists locally, if not download it
cog_filename = f"data/{Path(cog_href).name}"
if not os.path.isfile(cog_filename):
with requests.get(cog_href, stream=True) as r:
r.raise_for_status()
with open(cog_filename, "wb") as f:
for chunk in r.iter_content(chunk_size=8192):
f.write(chunk)
cog = rioxarray.open_rasterio(cog_filename, chunks=True)
print("==" * 10)
print("success")
breakThe next steps are to create a definitive mask using the amalgamation of the valid pixels layer and the cloud mask layer.
result = valid + cloud
# Set values greater than 1 to 0, others remain as 1
result = xr.where(result > 1, 0, 1)If we now multiply the cloud optimised geotif file by the result maskfile, expanded to match the shape of cog then each of the bands will have the clouds removed. This is a brute force approach for the purposes of generating a demonstrable workflow and in an operational context a user would likely want to undertand the impact of cloud in individual bands and use specific methods to remove that.
Finally the output is saved to a file
# Multiply cog.tif by the result
# We need to expand the result to match the shape of cog
fin = cog * result.squeeze("band").expand_dims(band=cog.band)
# save to a file
fin.rio.to_raster(raster_path=f"data/rm_cloud.tif", tiled=True, lock=threading.Lock())As mentioned, one method of generating the composite is to use pktools. It’s an older code, but it checks out. An alternative using Python might be to use xarray and the functions within that package. pktools works well for this use case as the command can be inserted directly into the workflow CWL. More information regarding this excellent tool can be found here:
https://pktools.nongnu.org/html/pkcomposite.html
Outputs
The following images provide an indication of how the pkcomposite tool works. The first image shows three input images (single band in this instance, coloured red, green and blue for clarity) that overlap a specific area of interest. The AOI is marked by the red boundary.
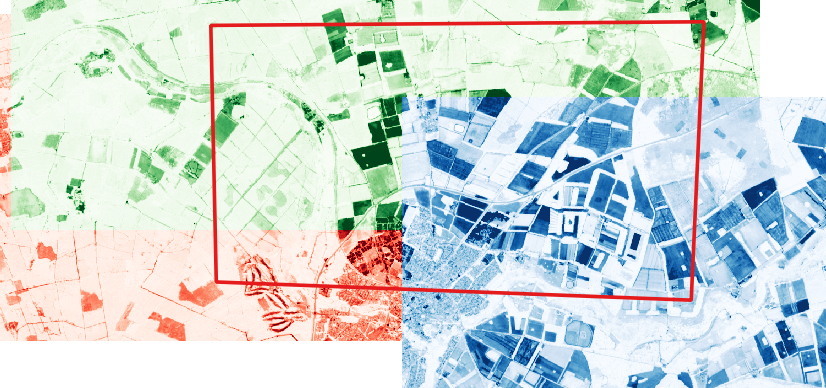
The second image plots the result of the pkcomposite command over the top. It shows how a median layer is created within the bounding box of the AOI.

In an EOAP cointext this could be scaled up across multiple multi-band images for a larger AOI.
Other methods
There are plenty of alternative ways in which the cloudless mosaic could be generated. One example is shown here for the Planetary Computer. It should be reasonably easy to transfer the procesisng chain explained in that example into something that would run in the JupyterHub instance on the EODH.
Generating the workflow
We can use eoap-gen to create a suitable workflow for this methodology. The first thing to do is understand the steps and the flow of data through the EODH workflow runner. The following diagram demonstrates how images are searched for and then scattered across numerous processing nodes to remove the cloud. The cloud-free images are then returned to fewer nodes to be mosaiced by month. Finally the mosaics are written out to a searchable STAC directory.
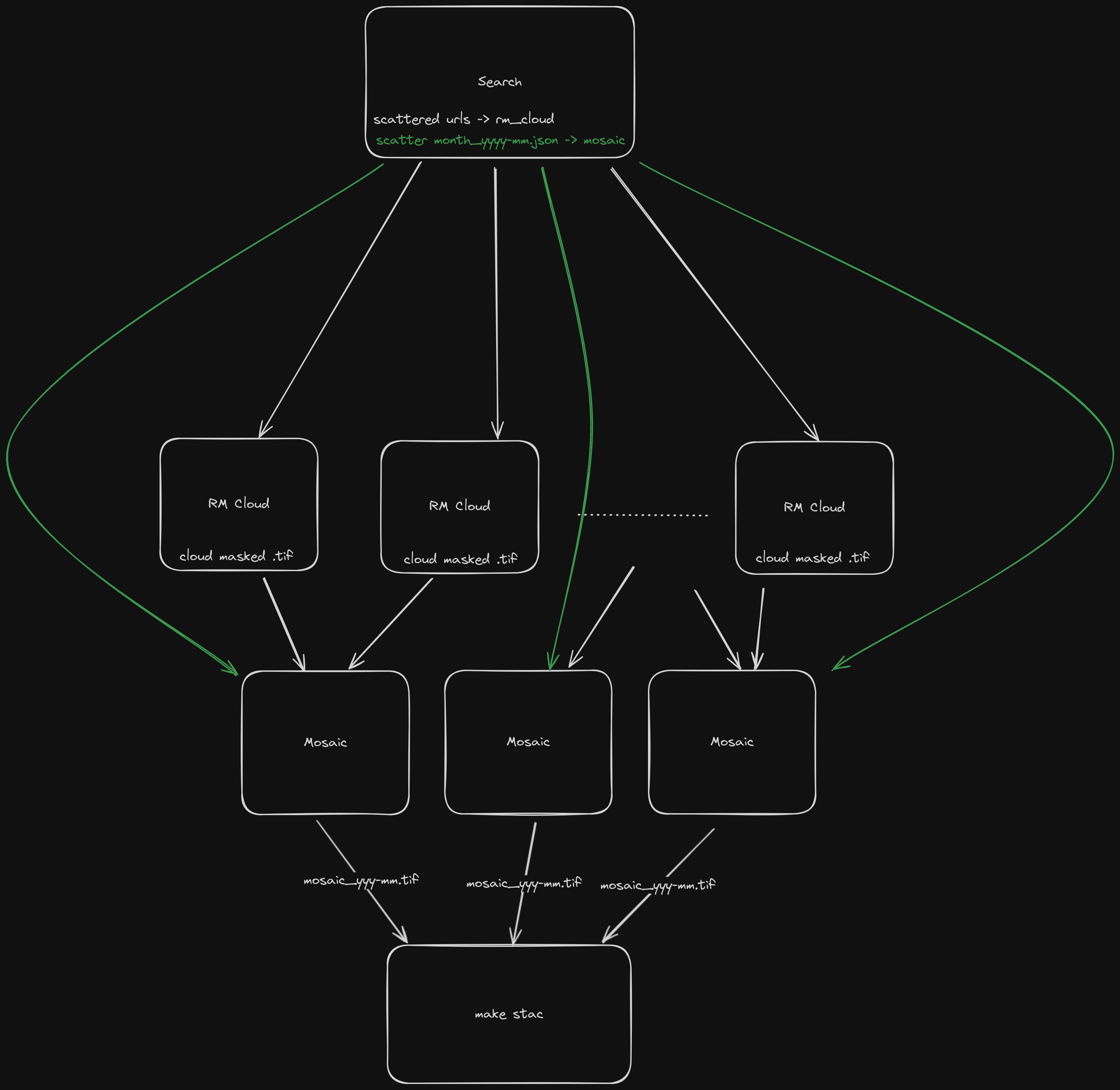
Using the configuration file here and reproduced below, it is possible to run eoap-gen to create the full package requirements.
id: cloud-free-best-pixel
doc: Generate cloud free best pixel mosaic on a per month basis
label: Cloud free best pixel
resources: # current ADES max is 4 cores and 16GB RAM, we need all we can get
cores_min: 4
ram_min: 16000
inputs:
- id: catalog
label: Catalog path
doc: Full catalog path
type: string
default: supported-datasets/ceda-stac-catalogue
- id: collection
label: collection id
doc: collection id
type: string
default: sentinel2_ard
- id: intersects
label: Intersects
doc: >
a GeoJSON-like json string, which provides a "type" member describing the type of the geometry and "coordinates"
member providing a list of coordinates. Will search for images intersecting this geometry.
type: string
default: >
{
"type": "Polygon",
"coordinates": [
[
[0.08905898091569497, 52.69722175598818],
[0.08905898091569497, 52.15527412683906],
[0.9565339502005088, 52.15527412683906],
[0.9565339502005088, 52.69722175598818],
[0.08905898091569497, 52.69722175598818]
]
]
}
- id: start_datetime
label: Start datetime
doc: Start datetime
type: string
default: "2023-04-01"
- id: end_datetime
label: End datetime
doc: End datetime
type: string
default: "2023-06-30"
outputs:
- id: stac_output
type: Directory
source: s2_make_stac/stac_catalog
steps:
- id: s2_search
script: S2-cloud-free-best-pixel/cli/search/search.py
requirements: S2-cloud-free-best-pixel/cli/search/requirements.txt
inputs:
- id: catalog
source: cloud-free-best-pixel/catalog
- id: collection
source: cloud-free-best-pixel/collection
- id: intersects
source: cloud-free-best-pixel/intersects
- id: start_datetime
source: cloud-free-best-pixel/start_datetime
- id: end_datetime
source: cloud-free-best-pixel/end_datetime
outputs:
- id: urls
type: string[]
outputBinding:
loadContents: true
glob: urls.txt
outputEval: $(self[0].contents.split('\n'))
- id: months
type: File[]
outputBinding:
glob: month_*.json
- id: s2_rm_cloud
script: S2-cloud-free-best-pixel/cli/rm_cloud/rm_cloud.py
requirements: S2-cloud-free-best-pixel/cli/rm_cloud/requirements.txt
conda:
- dask
- gdal
scatter_method: dotproduct
inputs:
- id: item_url
source: s2_search/urls
scatter: true
outputs:
- id: cloud_masked
type: File
outputBinding:
glob: "*.tif"
- id: s2_mosaic
script: S2-cloud-free-best-pixel/cli/mosaic/mosaic.py
requirements: S2-cloud-free-best-pixel/cli/mosaic/requirements.txt
apt_install:
- pktools
scatter_method: dotproduct
inputs:
- id: intersects
source: cloud-free-best-pixel/intersects
- id: month_json
source: s2_search/months
scatter: true
type: File
- id: all_images
source: s2_rm_cloud/cloud_masked
type: File[]
outputs:
- id: best_pixel
type: File
outputBinding:
glob: "*.tif"
- id: s2_make_stac
script: S2-cloud-free-best-pixel/cli/make_stac/make_stac.py
requirements: S2-cloud-free-best-pixel/cli/make_stac/requirements.txt
inputs:
- id: geometry
source: cloud-free-best-pixel/intersects
- id: files
source: s2_mosaic/best_pixel
type: File[]
outputs:
- id: stac_catalog
outputBinding:
glob: .
type: DirectoryRunning the configuration file results in the directory available here which can then be submitted to EODH using pyeodh and run using either the API client or QGIS plugin.